Deep Learning-Based Segmentation of Biological Networks in Fluorescence Microscopy
Introduction
Biological networks, such as endoplasmic reticulum (ER) and mitochondria (MITO) networks, are common in fluorescence microscopy. Accurate segmentation of these structures yields accurate shape representation which is crucial to quantitative analysis in many biomedical studies, such as drug screening and disease diagnosis. So far, however, there lacks a comprehensive study and a customized method for segmentation of such structures in fluorescence microscopy images (FLMI). In this project, we developed a deep learning-based pipeline to study the effects of image pre-processing, loss functions and model architectures for accurate segmentation of biological networks in FLMI.
Datasets
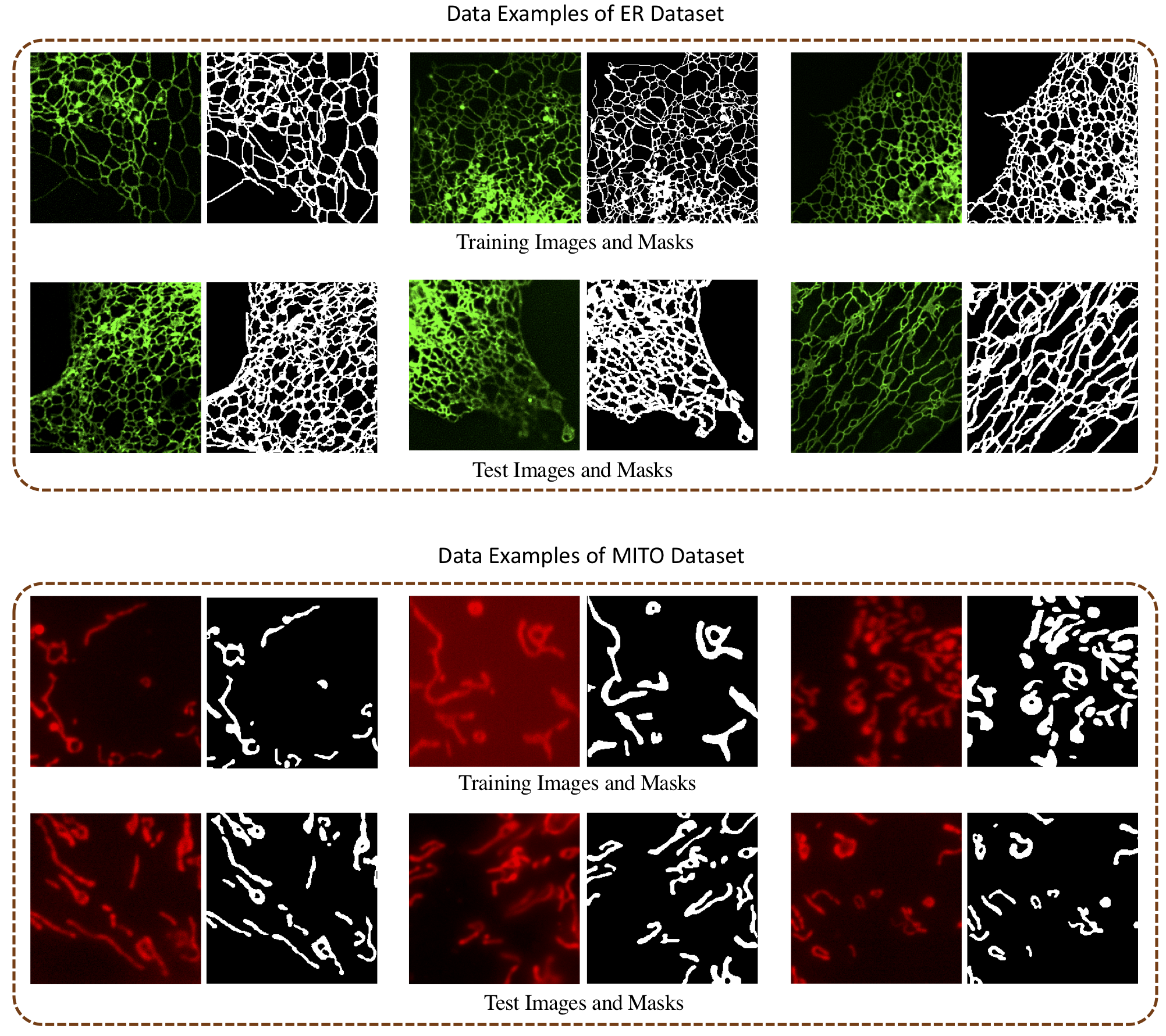
To support this study, we collect two datasets of typical FLMI of biological networks with manual mask annotations for ER and MITO. All images are cropped into 256x256 patches. To further increase the training size, we use horizontal, vertical flipping and 90°/180°/270° degrees rotation as augmentation which results in the final training set of MITO and ER with 1980 and 942 images respectively. For testing, we have 40 and 10 images respectively for the ER and MITO dataset. We provide both datasets for academic purposes. Please find the datasets from link.
Image examples
We show some image examples from our datasets in following figures.
Methodology
Please reference our paper for more technical details paper link. Corresponding codes can be found at github link.
Please cite our paper if you use our datasets:
@article{guo2020improvesegmentation, title={Improving Deep Learning-Based Segmentation of Biological Networks in Fluorescence Microscopy Using Simple Heuristics}, author={Yuanhao Guo, Yaoru Luo, Wenjing Li, Ge Yang}, journal={Submitted to journal}, year={2020}, }
Please contact us if you are interested in the project and want to collaborate: ge.yang@ia.ac.cn.