Research interests are in:
- computational and systems biology;
- biomedical image informatics.
Current research topics include:
- Computational analysis and modeling of spatiotemporal organization of intracellular organelle networks
- Computational analysis and modeling of organelle transport in complex geometry of neurons
- Deep learning based techniques for computational analysis of biological images
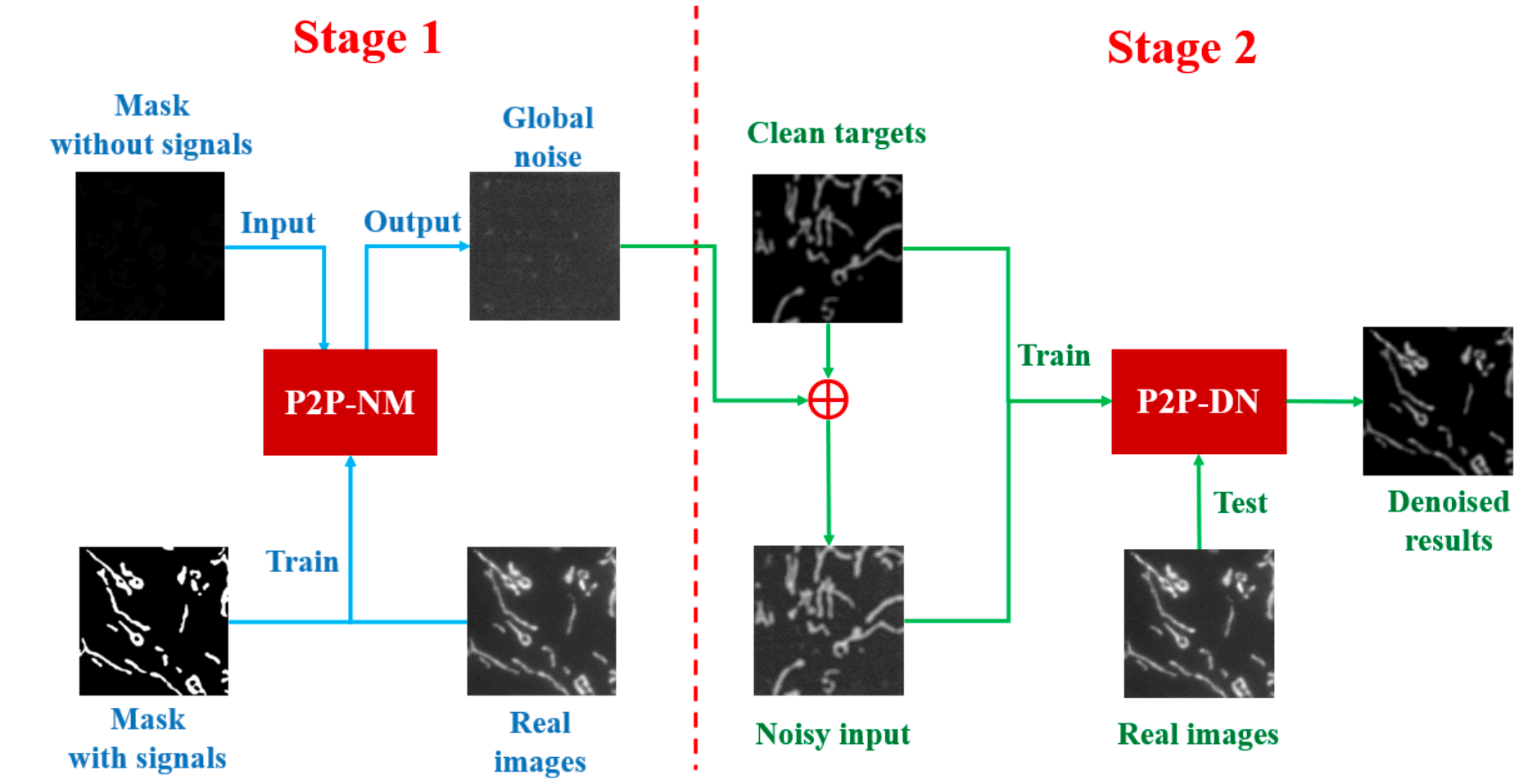
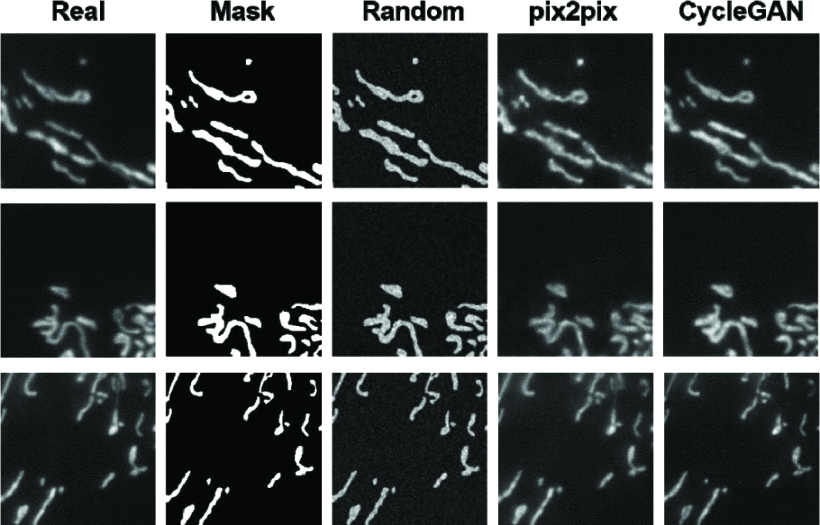
Blind Denoising of Fluorescence Microscopy Images Using GAN-Based Global Noise Modeling
We have developed a blind denoiser that uses one GAN to model image noise globally and another GAN to drastically reduce background noise.
Dynamic Organization of Intracellular Organelle Networks
We have developed a method to assess quality of synthetic fluorescence microscopy images and to evaluate their training performance in image segmentation.
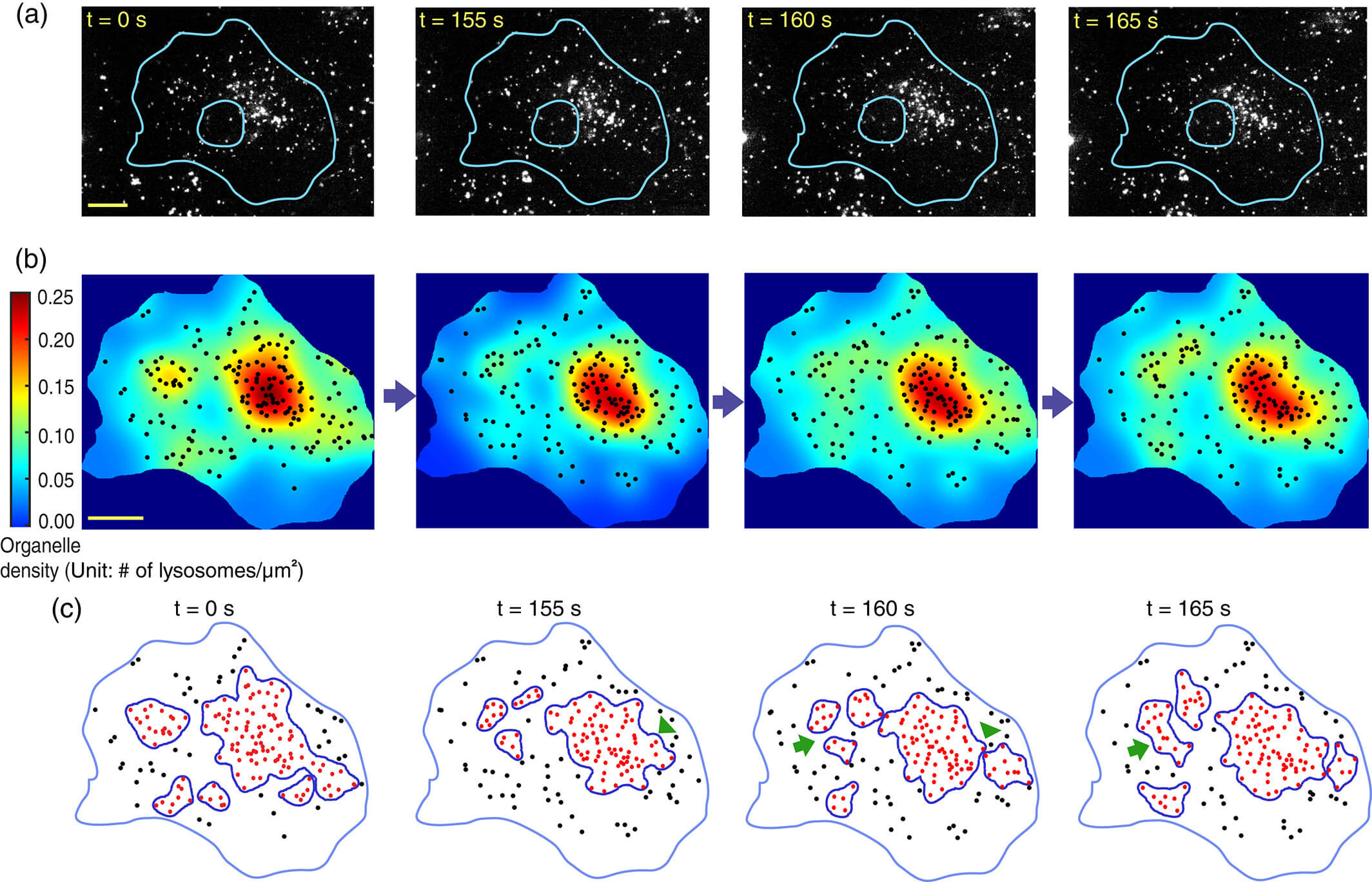
Whole-Cell Scale Dynamic Organization of Lysosomes Revealed by Spatial Statistical Analysis
Our findings reveal whole-cell scale spatial organization of lysosomes and provide insights into how organelle interactions are mediated and regulated across the entire intracellular space.
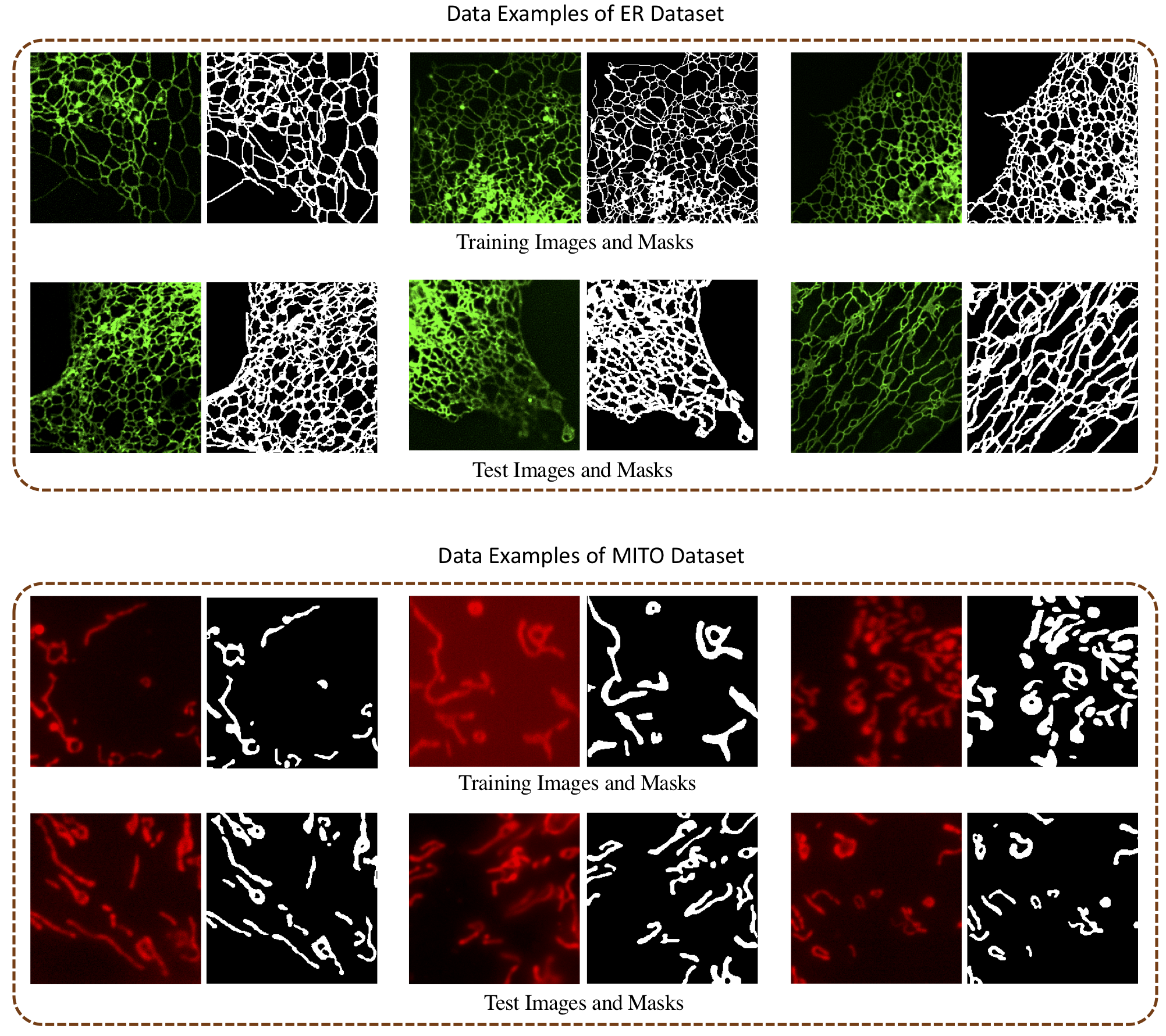
Deep Learning-Based Segmentation of Biological Networks in Fluorescence Microscopy
We developed a deep learning-based pipeline to study the effects of image pre-processing, loss functions and model architectures for accurate segmentation of biological networks in FLMI.